

Pseudomonas aeruginosa is a bacterial strain that can be responsible for several human diseases. The most serious include malignant external otitis, endophthalmitis, endocarditis, meningitis, pneumonia, and septicemia.
The environments in which these bacteria are most frequently found include soil, plants, and water. They can even be found on human and animal skin, without causing illness, in a process known as bacterial colonization. Microbiological research can help establish the cause of certain infectious diseases, making it easier to choose the best treatment. This is why it is important to find a quick and easy way to identify these bacteria.
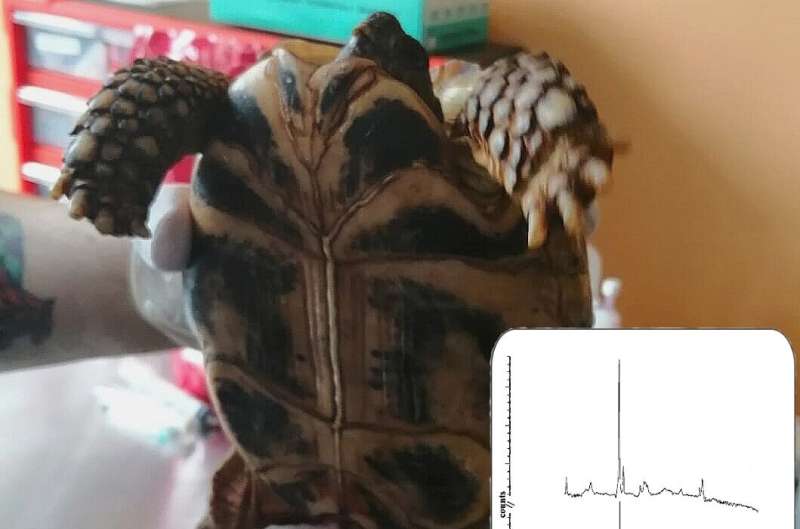
A new study, published in BioRisk, has explored this by applying spectroscopic techniques for quick analysis directly from an object, which in this case was turtle skin.
“Microbial organisms play key roles in animal health and ecology. The European pond turtle often lives in city zoo gardens and private houses. Often, the most commonly found bacteria from turtle skin surfaces was Pseudomonas species,” says Aleksandrs Petjukevics of Daugavpils University, whose team conducted the study.
What is Raman spectroscopy?
“Classical microbiological research techniques have several disadvantages: first of all, it is a rather lengthy process. The minimum period is 3-4 days, but many days and even weeks may pass before the isolated pathogen is accurately identified, and it uses expensive chemicals and resources,” says Petjukevics.
As an alternative, spectrometry makes it possible to identify a prepared sample of a microorganism while reducing the identification time to 5-30 minutes. Raman spectra represent an ensemble of signals that arise from the molecular vibrations of individual cell components of gram-negative bacteria, integrating over proteins, lipids, and carbohydrates.
“This non-destructive chemical analysis technique provides detailed information about chemical structure, phase and polymorphy, crystallinity, and molecular interactions. It is based on the interaction of light with the chemical bonds within a material,” Petjukevics says.
Research results and implications
The study’s findings showed that Pseudomonas bacteria can be quickly identified using this detection technology, with excellent analytical and diagnostic sensitivity, making it a dependable technique. Unlike other methods, this technique does not require long-term bacterial sample preparation and expensive reagents, which makes it promising for studying other strains of bacteria.
“This study demonstrated the ability to obtain fast and high-quality Raman spectra of bacterial cells using vibrational spectroscopy,” concludes Petjukevics. “Raman spectroscopy can be considered an express method for identifying microorganisms. It holds great potential for future research involving different microorganisms.”
More information:
Aleksandrs Petjukevičs et al, Prospects and possibilities of using Raman spectroscopy for the identification of Pseudomonas aeruginosa from turtle Emys orbicularis (Linnaeus, 1758) skin, BioRisk (2023). DOI: 10.3897/biorisk.21.111983
Provided by
Pensoft Publishers
Citation:
Novel bacteria identification methods might help speed up disease diagnosis (2023, December 14)
retrieved 14 December 2023
from https://phys.org/news/2023-12-bacteria-identification-methods-disease-diagnosis.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.





